DETERMINING THE OCCURRENCE OF SOME VIRULENCE GENES IN PROTEUS SPECIES ISOLATES
Views: 223 https://doi.org/10.59807/jlsar.v4i2.88
https://doi.org/10.59807/jlsar.v4i2.88
Keywords:
Proteus species, Urinary tract infections, Extended-spectrum β-lactamases, polymerase chain reaction, Virulence genesAbstract
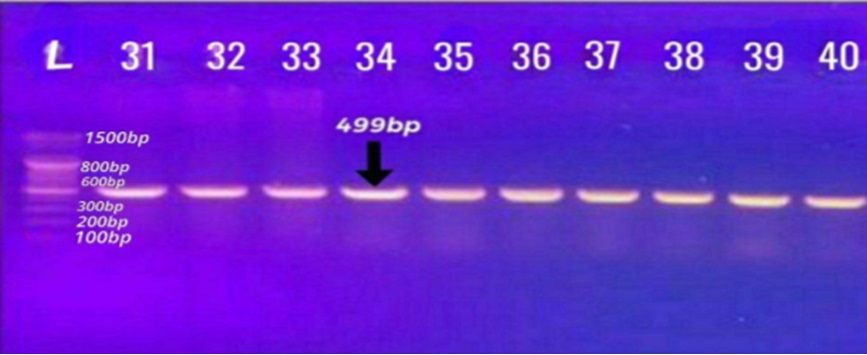
Forty isolates of Gram-negative rod-shaped bacteria termed as Proteus, widely known for their swarming motility and urease activity, which cause complicated urinary tract infections (UTIs), were isolated and identified. Two hundred and ten urine specimens collected from the patients suffering from UTIs, who were hospitalized in Babylon hospitals, were used for the isolation of Proteus species isolates. The morphological features (for cells and colonies), biochemical tests, VITEK 2 compact and polymerase chain reaction (PCR) for Proteus-specific genes were used for the identification of these isolates. The assessment of the antimicrobial profiles that represent the prevalence and the level of pathogenicity of the isolates was also carried out. Furthermore, the whole genomic DNA of the isolates was extracted to determine the sites of blaTEM, blaCTX-M, and blaSHV genes on the genome of the isolates. The results revealed that thirty isolates were P. mirabilis and ten isolates were P. vulgaris. These isolates were given names as PM1 to PM30 for P. mirabilis and PV31 to PV40 for P. vulgaris. The most effective antibiotics against the isolates were erythromycin (97.5%), followed by tobramycin (85%), ampicillin (82.5%), chloramphenicol (60%), piperacillin (55%) and 52.5% for each sulfamethoxazole and azithromycin. The meropenem and imipenem showed less resistance (35%) followed by ciprofloxacin (30%) and gentamicin (15%). The PCR assay exhibited that these isolates carried blaTEM gene at the rate of 38/40 (95%), blaSHV gene at the rate of 33/40 (82.5%) and blaCTX-M gene at the rate of 37/40 (92.5%). Therefore, reducing the frequency and severity of infections, however, more research is needed to understand how the rates of pathogenicity of Proteus species isolates can be controlled.
Downloads
References
J. E. Bennett, R. Dolin, and M. J. Blaser, Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, vol. 1–2. 2014. doi: 10.1016/s1473-3099(10)70089-x.
J. N. Schaffer and M. M. Pearson, “ Proteus mirabilis and Urinary Tract Infections ,” Microbiol Spectr, vol. 3, no. 5, 2015, doi: 10.1128/microbiolspec.uti-0017-2013.
B. N. Kim, N. J. Kim, M. N. Kim, Y. S. Kim, J. H. Woo, and J. Ryu, “Bacteraemia due to tribe proteeae: A review of 132 cases during a decade (1991-2000),” Scand J Infect Dis, vol. 35, no. 2, 2003, doi: 10.1080/0036554021000027015.
J. Bloch, X. Lemaire, L. Legout, D. Ferriby, Y. Yazdanpanah, and E. Senneville, “Brain abscesses during Proteus vulgaris bacteremia,” Neurological Sciences, vol. 32, no. 4, 2011, doi: 10.1007/s10072-010-0408-0.
C. E. Armbruster, H. L. T. Mobley, and M. M. Pearson, “ Pathogenesis of Proteus mirabilis In-fection ,” EcoSal Plus, vol. 8, no. 1, 2018, doi: 10.1128/ecosalplus.esp-0009-2017.
J. Kwiecińska-Piróg, K. Skowron, and E. Gospodarek-Komkowska, “Primary and secondary bacteremia caused by Proteus spp.: Epidemiology, strains susceptibility and biofilm formation,” Pol J Microbiol, vol. 67, no. 4, 2018, doi: 10.21307/pjm-2018-055.
A. Wang, J. G. Gaca, and V. H. Chu, “Management considerations in infective endocarditis: A review,” JAMA - Journal of the American Medical Association, vol. 320, no. 1. 2018. doi: 10.1001/jama.2018.7596.
L. M. Baddour et al., “Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications,” Circulation, vol. 132, no. 15, 2015.
S. Minami, A. Yotsuji, M. Inoue, and S. Mitsuhashi, “Induction of β-lactamase by various β-lactam antibiotics in Enterobacter cloacae,” Antimicrob Agents Chemother, vol. 18, no. 3, 1980, doi: 10.1128/AAC.18.3.382.
C. Mugnaioli et al., “CTX-M-type extended-spectrum β-lactamases in Italy: Molecular epide-miology of an emerging countrywide problem,” Antimicrobial Agents and Chemotherapy, vol. 50, no. 8. 2006. doi: 10.1128/AAC.00068-06.
F. Odds, “Biochemical Tests for Identification of Medical Bacteria,” J Clin Pathol, vol. 34, no. 5, 1981, doi: 10.1136/jcp.34.5.572-a.
Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Suscepti-bility Testing:CLSI supplement M100, vol. 8, no. 3. 2023.
M. Bokaeian, S. S. Zahedani, M. S. Bajgiran, and A. A. Moghaddam, “Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa Producing Ex-tended Spectrum β-lactamases,” Jundishapur J Microbiol, vol. 8, no. 1, 2015, doi: 10.5812/jjm.13783.
S. P. Lin, M. F. Liu, C. F. Lin, and Z. Y. Shi, “Phenotypic detection and polymerase chain reac-tion screening of extended-spectrum β-lactamases produced by Pseudomonas aeruginosa iso-lates,” Journal of Microbiology, Immunology and Infection, vol. 45, no. 3, 2012, doi: 10.1016/j.jmii.2011.11.015.
A. Mirzaei, B. Nasr Esfahani, A. Raz, M. Ghanadian, and S. Moghim, “From the Urinary Cath-eter to the Prevalence of Three Classes of Integrons, β -Lactamase Genes, and Differences in Antimicrobial Susceptibility of Proteus mirabilis and Clonal Relatedness with Rep-PCR,” Bio-med Res Int, vol. 2021, 2021, doi: 10.1155/2021/9952769.
T. Hussain et al., “Uropathogens Antimicrobial Sensitivity and Resistance Pattern From Out-patients in Balochistan, Pakistan,” Cureus, 2021, doi: 10.7759/cureus.17527.
C. Bourély et al., “Antimicrobial resistance patterns of bacteria isolated from dogs with otitis,” Epidemiol Infect, vol. 147, 2019, doi: 10.1017/S0950268818003278.
R. Cantón et al., “Monitoring the antimicrobial susceptibility of gram-negative organisms in-volved in intraabdominal and urinary tract infections recovered during the smart study (Spain, 2016 and 2017),” Revista Espanola de Quimioterapia, vol. 32, no. 2, 2019.
S. Bilal et al., “Antimicrobial profiling and molecular characterization of antibiotic resistant genes of Proteus vulgaris isolated from tertiary care hospital, Islamabad, Pakistan,” Pak J Pharm Sci, vol. 32, no. 6, 2019.
A. M. Lazm, M. Sh. Jebur, and G. B. Alomashi, “Sequencing of hpmA gene in Proteus mirabilis of UTIs among rheumatoid arthritis patients,” Journal of Pharmaceutical Sciences and Research, vol. 10, no. 2, 2018.
N. Fam, “Prevalence of Plasmid-Mediated ampC Genes in Clinical Isolates of Enterobacteri-aceae from Cairo, Egypt,” Br Microbiol Res J, vol. 3, no. 4, 2013, doi: 10.9734/bmrj/2013/4653.
O. Daini, M. Effiong, and O. Ogbolu, “Quinolones Resistance And R-Plasmids Of Clinical Iso-lates Of Pseudomonas Species,” Sudan Journal of Medical Sciences, vol. 3, no. 2, 2008, doi: 10.4314/sjms.v3i2.38528.
D. N. Al Obeidi, Z. M. Alzubaidy, and A. Ammari, “Molecular detection of some virulence factors of uropathogenic proteus mirabilis using PCR technique,” in AIP Conference Proceedings, 2023. doi: 10.1063/5.0103334.
R. Cantón and T. M. Coque, “The CTX-M β-lactamase pandemic,” Current Opinion in Microbi-ology, vol. 9, no. 5. 2006. doi: 10.1016/j.mib.2006.08.011.
Y. Ishii, A. Ohno, H. Taguchi, S. Imajo, M. Ishiguro, and H. Matsuzawa, “Cloning and se-quence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli,” Antimicrob Agents Chemother, vol. 39, no. 10, 1995, doi: 10.1128/AAC.39.10.2269.
N. Shibata et al., “PCR classification of CTX-M-type β-lactamase genes identified in clinically isolated gram-negative bacilli in Japan,” Antimicrob Agents Chemother, vol. 50, no. 2, 2006, doi: 10.1128/AAC.50.2.791-795.2006.
S. Uzunović, A. Ibrahimagić, and B. Bedenić, “AUTHOR’S CORRECTION: Molecular epide-miology and antimicrobial susceptibility of AmpC- and/or extended-spectrum (ESBL) ß-lactamaseproducing Proteus spp. clinical isolates in Zenica-Doboj Canton, Bosnia and Her-zegovina,” Med Glas (Zenica), vol. 14, no. 2, 2017.
A. Malaki, E. Ferdosi-Shahandashti, A. Maali, P. Sabbagh, and A. Khademian, “Molecular Characterizations and Antimicrobial Susceptibility of Extended-Spectrum ß-Lactamase (ESBL) Producing Proteus spp. Clinical Isolates in Babol, Northern Iran,” Journal of Medical Microbi-ology and Infectious Diseases, vol. 10, no. 3, pp. 114–121, 2022.
G. Rajivgandhi, M. Maruthupandy, and N. Manoharan, “Detection of TEM and CTX-M genes from ciprofloxacin resistant Proteus mirabilis and Escherichia coli isolated on urinary tract infections (UTIs),” Microb Pathog, vol. 121, 2018, doi: 10.1016/j.micpath.2018.05.024.
P. A. Bradford, “Extended-spectrum β-lactamases in the 21st century: Characterization, epi-demiology, and detection of this important resistance threat,” Clinical Microbiology Reviews, vol. 14, no. 4. 2001. doi: 10.1128/CMR.14.4.933-951.2001.
X. Qin et al., “Prevalence and mechanisms of broad-spectrum β-lactam resistance in Entero-bacteriaceae: A children’s hospital experience,” Antimicrob Agents Chemother, vol. 52, no. 11, 2008, doi: 10.1128/AAC.00622-08.
T. M. Coque et al., “Dissemination of clonally related Escherichia coli strains expressing ex-tended-spectrum β-lactamase CTX-M-15,” Emerg Infect Dis, vol. 14, no. 2, 2008, doi: 10.3201/eid1402.070350.
J. K. Bailey, J. L. Pinyon, S. Anantham, and R. M. Hall, “Distribution of the bla TEM Gene and bla TEM-Containing Transposons in Commensal Escherichia coli,” Journal of Antimicrobial Chemotherapy, vol. 8, no. 6, pp. 745–751, 2011.
M. Shaaban, O. A. A. El-Rahman, B. Al-Qaidi, and H. M. Ashour, “Antimicrobial and antibio-film activities of probiotic lactobacilli on antibiotic-resistant proteus mirabilis,” Microorgan-isms, vol. 8, no. 6, 2020, doi: 10.3390/microorganisms8060960.
S. Gomaa, F. Serry, H. Abdellatif, and H. Abbas, “Elimination of multidrug-resistant Proteus mirabilis biofilms using bacteriophages,” Arch Virol, vol. 164, no. 9, 2019, doi: 10.1007/s00705-019-04305-x.
Published
How to Cite
Issue
Section
Citations
License
Copyright (c) 2023 Copyright (c) 2022 Creative Commons Attribution 4.0 International (CC-BY 4.0)

This work is licensed under a Creative Commons Attribution 4.0 International License.
This journal is licensed under a Creative Commons Attribution 4.0 International (CC-BY 4.0)