GENETIC DIVERSITY AND ANTIBIOTIC RESISTANCE PATTERNS OF PSEUDOMONAS AERUGINOSA ISOLATES FROM IRAQI HOSPITALS
Views: 203 https://doi.org/10.59807/jlsar.v5i1.93
https://doi.org/10.59807/jlsar.v5i1.93
Keywords:
P. aeruginosa, Multidrug-Resistance, DNA Sequencing, Virulence FactorsAbstract
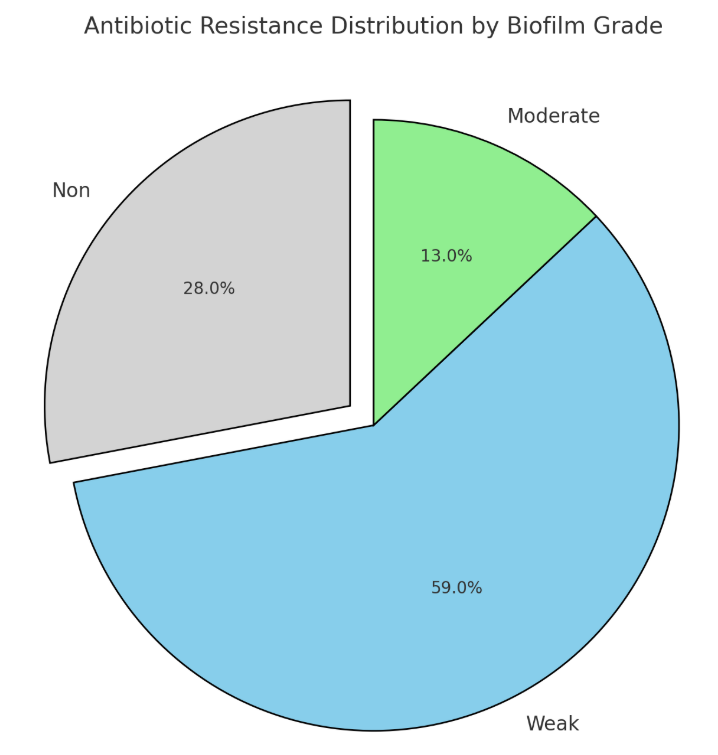
Forty-six isolates of Pseudomonas aeruginosa were identified from 150 samples taken from different patients admitted to general hospitals in nine cities of Iraq during the period from September 2021 to January 2022. The isolates were named PA1 to PA46. The biofilm formation by these isolates was studied using microtiter plate assays, and the correlation between biofilm grades and antibiotic resistance was also analyzed. It was found that 34 isolates were able to produce biofilm. To determine the locations of blaTEM, blaCTX-M, and blaSHV genes on the genomes of the isolates, the whole genomic DNA of nine isolates, PA1 to PA9, was extracted, and the sequencing of these genes was achieved. Matching the isolates with NCBI GenBank global Pseudomonas strains showed that one isolate (PA1) was related to the UAE, two isolates (PA2 and PA3) were related to India, three isolates (PA4, PA5, and PA6) were related to Egypt, and three isolates (PA7, PA8, and PA9) were related to Iran. Hence, the variable frequencies in the sequencing of blaTEM, blaCTX-M, and blaSHV genes need further studies to create a genetic diversity map of P. aeruginosa.
Downloads
References
M. A. A. Mohammed Saleh and H. F. Naji, “Detection of blatem, blactx-m, and blashv genes in clinical isolates of multidrug-resistant pseudomonas aeruginosa,” Int J Health Sci (Qassim), 2022, doi: 10.53730/ijhs.v6ns7.12451.
F. Stapleton and N. Carnt, “Contact lens-related microbial keratitis: How have epidemiology and genetics helped us with pathogenesis and prophylaxis,” in Eye, 2012. doi: 10.1038/eye.2011.288.
T. G. Thuo, C. Kiyuukia, J. Maina, T. Judah, S. Kiiru, and J. Kiiru, “Antimicrobial Resistance Profiles and Clonal Relatedness of <i>Pseudomonas aeruginosa</i> Strains Recov-ered from Wounds Infections of Outpatient Population Presenting in a Rural Hospital in Kenya,” Adv Microbiol, vol. 09, no. 02, 2019, doi: 10.4236/aim.2019.92009.
L. Hall-Stoodley, J. W. Costerton, and P. Stoodley, “Bacterial biofilms: From the natural envi-ronment to infectious diseases,” 2004. doi: 10.1038/nrmicro821.
T. K. W. Ling, J. Xiong, Y. Yu, C. C. Lee, H. Ye, and P. M. Hawkey, “Multicenter antimicrobial susceptibility survey of gram-negative bacteria isolated from patients with communi-ty-acquired infections in the People’s Republic of China,” Antimicrob Agents Chemother, vol. 50, no. 1, 2006, doi: 10.1128/AAC.50.1.374-378.2006.
M. Nasser, S. Gayen, and A. S. Kharat, “Prevalence of β-lactamase and antibiotic-resistant Pseudomonas aeruginosa in the Arab region,” 2020. doi: 10.1016/j.jgar.2020.01.011.
K. Tamura, J. Dudley, M. Nei, and S. Kumar, “MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0,” Mol Biol Evol, vol. 24, no. 8, 2007, doi: 10.1093/molbev/msm092.
A. N. Suwaidan and H. F. Naji, “bla OXA genotyping of multidrug resistant Pseudomonas ae-ruginosa isolated from clinical specimens,” Eurasian J Biosci, vol. 14, no. 2, 2020.
S. P. Lin, M. F. Liu, C. F. Lin, and Z. Y. Shi, “Phenotypic detection and polymerase chain reac-tion screening of extended-spectrum β-lactamases produced by Pseudomonas aeruginosa iso-lates,” Journal of Microbiology, Immunology and Infection, vol. 45, no. 3, 2012, doi: 10.1016/j.jmii.2011.11.015.
N. Saitou and M. Nei, “The neighbor-joining method: a new method for reconstructing phylo-genetic trees.,” Mol Biol Evol, vol. 4, no. 4, 1987, doi: 10.1093/oxfordjournals.molbev.a040454.
D. Penny, “Inferring Phylogenies.—Joseph Felsenstein. 2003. Sinauer Associates, Sunderland, Massachusetts.,” Syst Biol, vol. 53, no. 4, 2004, doi: 10.1080/10635150490468530.
F. Odds, “Biochemical Tests for Identification of Medical Bacteria,” J Clin Pathol, vol. 34, no. 5, 1981, doi: 10.1136/jcp.34.5.572-a.
T. S. Murray, M. Ledizet, and B. I. Kazmierczak, “Swarming motility, secretion of type 3 effec-tors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeru-ginosa clinical isolates,” J Med Microbiol, vol. 59, no. 5, 2010, doi: 10.1099/jmm.0.017715-0.
J. Fricks-Lima et al., “Differences in biofilm formation and antimicrobial resistance of Pseu-domonas aeruginosa isolated from airways of mechanically ventilated patients and cystic fi-brosis patients,” Int J Antimicrob Agents, vol. 37, no. 4, 2011, doi: 10.1016/j.ijantimicag.2010.12.017.
S. Kumar, M. Nei, J. Dudley, and K. Tamura, “MEGA: A biologist-centric software for evolu-tionary analysis of DNA and protein sequences,” Brief Bioinform, vol. 9, no. 4, 2008, doi: 10.1093/bib/bbn017.
Published
How to Cite
Issue
Section
Citations
License
Copyright (c) 2024 Copyright (c) 2024 Creative Commons Attribution 4.0 International (CC-BY 4.0)

This work is licensed under a Creative Commons Attribution 4.0 International License.
This journal is licensed under a Creative Commons Attribution 4.0 International (CC-BY 4.0)